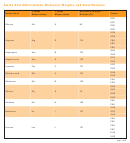
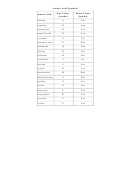
Properties Of Amino Acids Chart
ADVERTISEMENT
Properties of amino acids
Occurrence
Accessible
Ranking of
Amino acid
pK
of ionizing
Average residue
Monoisotopic
in proteins
c
Percent buried
V
e
van der Waals surface
amino acid
a
r
residue
side chain
a
mass
b
(daltons)
mass (daltons)
b
(%)
residues
d
(%)
(Å
3
)
volume
f
(Å
3
)
area
g
(Å
2
)
polarities
h
Alanine
–
71.0788
71.03711
7.5
38 (12)
92
67
67
9 (7)
Arginine
12.5 (>12)
156.1876
156.10111
5.2
0
225
148
196
15 (19)
Asparagine
–
114.1039
114.04293
4.6
10 (2)
135
96
113
16 (16)
Aspartic acid
3.9 (4.4–4.6)
115.0886
115.02694
5.2
14.5 (3)
125
91
106
19 (18)
Cysteine
8.3 (8.5–8.8)
103.1448
103.00919
1.8
47 (3)
106
86
104
7 (8)
Glutamine
–
128.1308
128.05858
4.1
6.3 (2.2)
161
114
144
17 (14)
Glutamic acid
4.3 (4.4–4.6)
129.1155
129.04259
6.3
20 (2)
155
109
138
18 (17)
Glycine
–
57.0520
57.02146
7.1
37 (10)
66
48
11 (9)
Histidine
6.0 (6.5–7.0)
137.1412
137.05891
2.2
19 (1.2)
167
118
151
10 (13)
Isoleucine
–
113.1595
113.08406
5.5
65 (12)
169
124
140
1 (2)
Leucine
–
113.1595
113.08406
9.1
41 (10)
168
124
137
3 (1)
Lysine
10.8 (10.0–10.2)
128.1742
128.09496
5.8
4.2 (0.1)
171
135
167
20 (15)
Methionine
–
131.1986
131.04049
2.8
50 (2)
171
124
160
5 (5)
Phenylalanine
–
147.1766
147.06841
3.9
48 (5)
203
135
175
2 (4)
Proline
–
97.1167
97.05276
5.1
24 (3)
129
90
105
13 (–)
Serine
–
87.0782
87.03203
7.4
24 (8)
99
73
80
14 (12)
Threonine
–
101.1051
101.04768
6.0
25 (5.5)
122
93
102
12 (11)
Tryptophan
–
186.2133
186.07931
1.3
23 (1.5)
240
163
217
6 (6)
Tyrosine
10.9 (9.6–10.0)
163.1760
163.06333
3.3
13 (2.2)
203
141
187
8 (10)
Valine
–
99.1326
99.06841
6.5
56 (15)
142
105
117
4 (3)
a
The pK
values in most cases are at 25ºC. The expected pK
values in proteins, shown in parentheses, are determined from model compounds in which titration of side chains is decoupled from charge
a
a
effects of α-substituents. (Data from Cantor and Schimmel 1980.)
b
Data from Burlingame and Carr (1996).
c
Frequency of occurrence of each amino acid residue in the primary structures of 105,990 sequences in the nonredundant OWL protein database (release 26.0 e) (Trinquier and Sanejouand 1998).
d
This column represents the tendency of an amino acid to be buried (defined as <5% of residue available to solvent) in the interior of a protein and is based on the structures of nine proteins (total
of ~2000 individual residues studied, with 587 [29%] of these buried). Values indicate how often each amino acid was found buried, relative to the total number of residues of this amino acid found in
the proteins (values in parentheses indicate the number of buried residues of this amino acid found relative to all buried residues in the proteins). (Data from Schien 1990; for other calculation meth-
ods with similar results, see Janin 1979 and Rose at al. 1985.)
e
Average volume (V
) of buried residues, calculated from the surface area of the side chain (Richards 1977; Baumann et al. 1989).
r
f
Data from Darby and Creighton (1993).
g
Total accessible surface area (ASA) of amino acid side chain for residue X in a Gly-X-Gly tripeptide with the main chain in an extended conformation (Miller et al. 1987). The ASA or cavity surface
area is defined as the surface traced by the center of a sphere with the radius of a water molecule (0.15 mm) as it is rolled over the surface of a molecular model of the solution (Lee and Richards 1971).
h
Values shown represent the mean ranking of amino acids according to the frequency of their occurrence at each sequence rank for 38 published hydrophobicity scales (Trinquier and Sanejouand
1998). Although the majority of these hydrophobicity scales are derived from experimental measurements of chemical behavior or physicochemical properties (e.g., solubility in water, partition between
water and organic solvent, chromatographic migration, or effects on surface tension) of isolated amino acids, several “operational” hydrophobicity scales based on the known environment characteris-
tics of amino acids in proteins, such as their solvent accessibility or their inclination to occupy the core of proteins (based on the position of residues in the tertiary structures as observed by X-ray crys-
tallography or NMR) are included (Trinquier and Sanejouand 1998). The lower rankings represent the most hydrophobic amino acids, and higher values represent the most hydrophilic amino acids. For
comparative purposes, the hydrophobicity scale of Radzicka and Wolfenden is shown in parentheses. This scale was derived from the measured hydration potential of amino acids that is based on their
free energies of transfer from the vapor phase to cyclohexane, 1-octanol, and neutral aqueous solution (Radzicka and Wolfenden 1988).
ADVERTISEMENT
0 votes
Related Articles
Related forms
Related Categories
Parent category: Education
 1
1